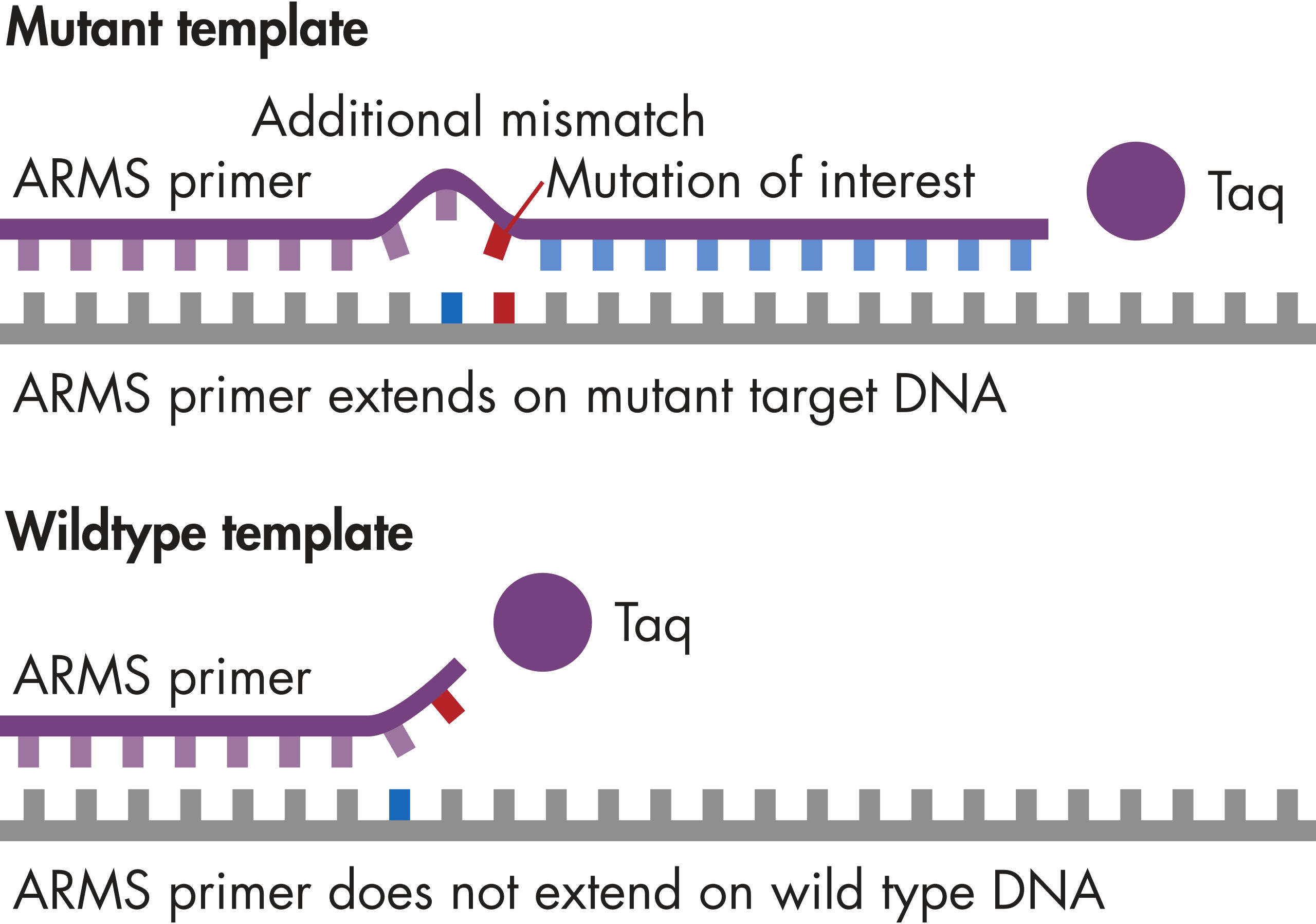
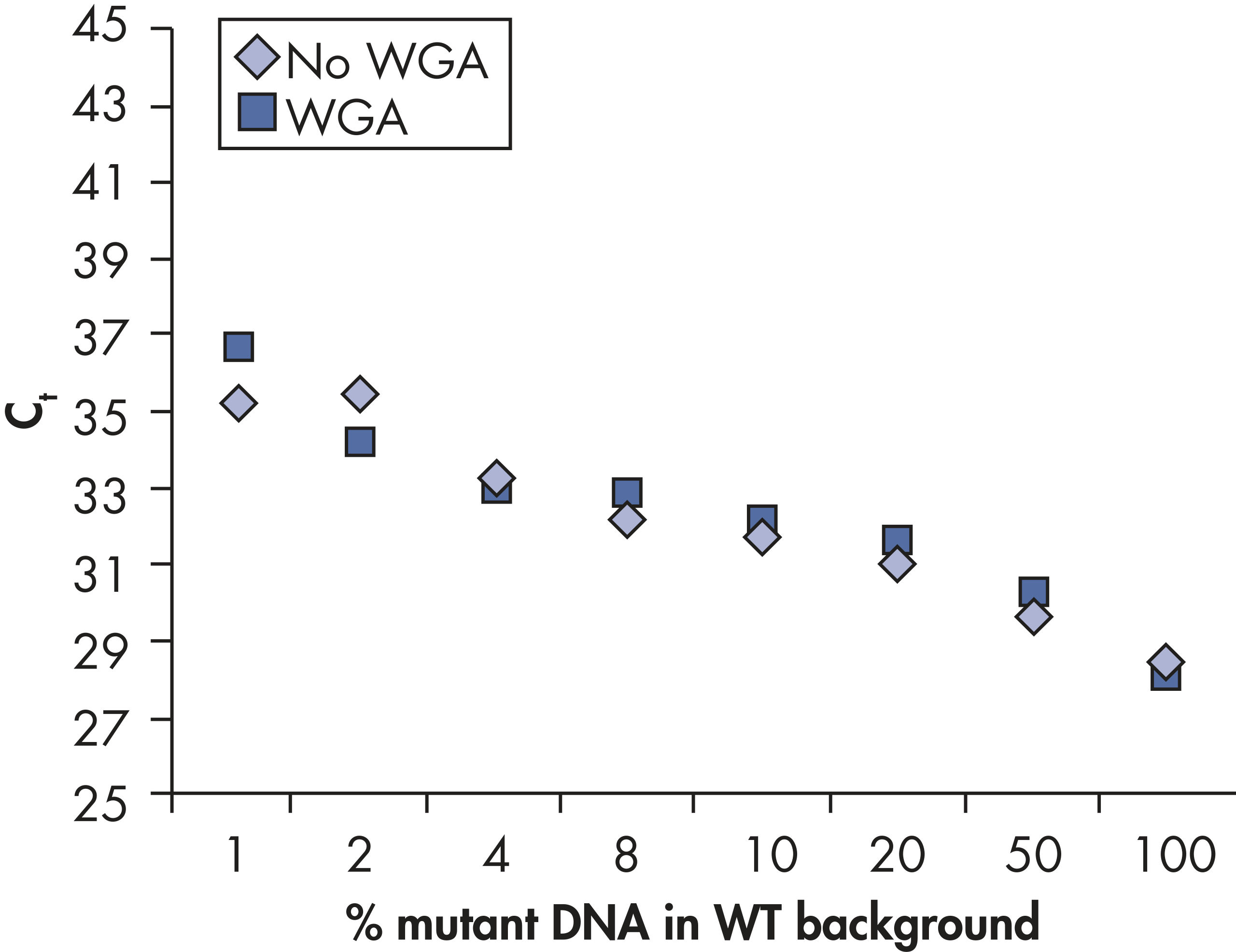
PCR plate and master mix
Target List
AKT1
The best known AKT1 mutations are c.49G>A, p.E17K, a PH domain mutation that results in constitutive targeting of AKT1 to plasma membrane, and an activating mutation, c.145G>A, p.E49K.
BRAF
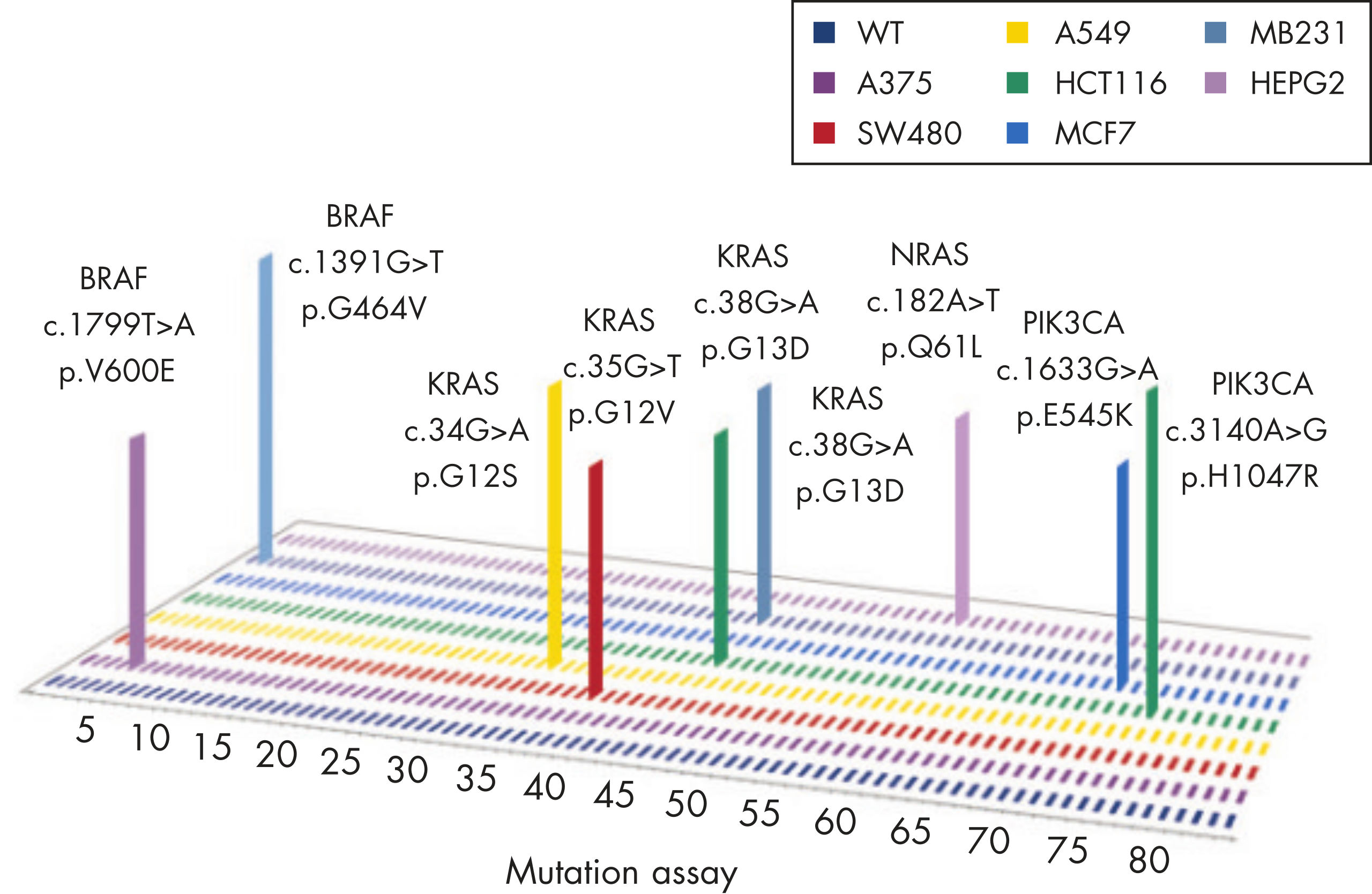
Common point mutations in BRAF either increase kinase activity, such as L597V, L597Q and V600E, or inhibit kinase activity, such as G464V, G466V and G469A.
CDKN2A
The most important CDKN2A loss-of-function mutations occur in the consensus ankyrin domain, which lead to the inability to form stable complexes with its targets.
CTNNB1
The most frequently detected CTNNB1 mutations result in abnormal WNT signaling activity. The mutated codons are mainly several serines and threonines targeted for phosphorylation by GSK3B.
EGFR
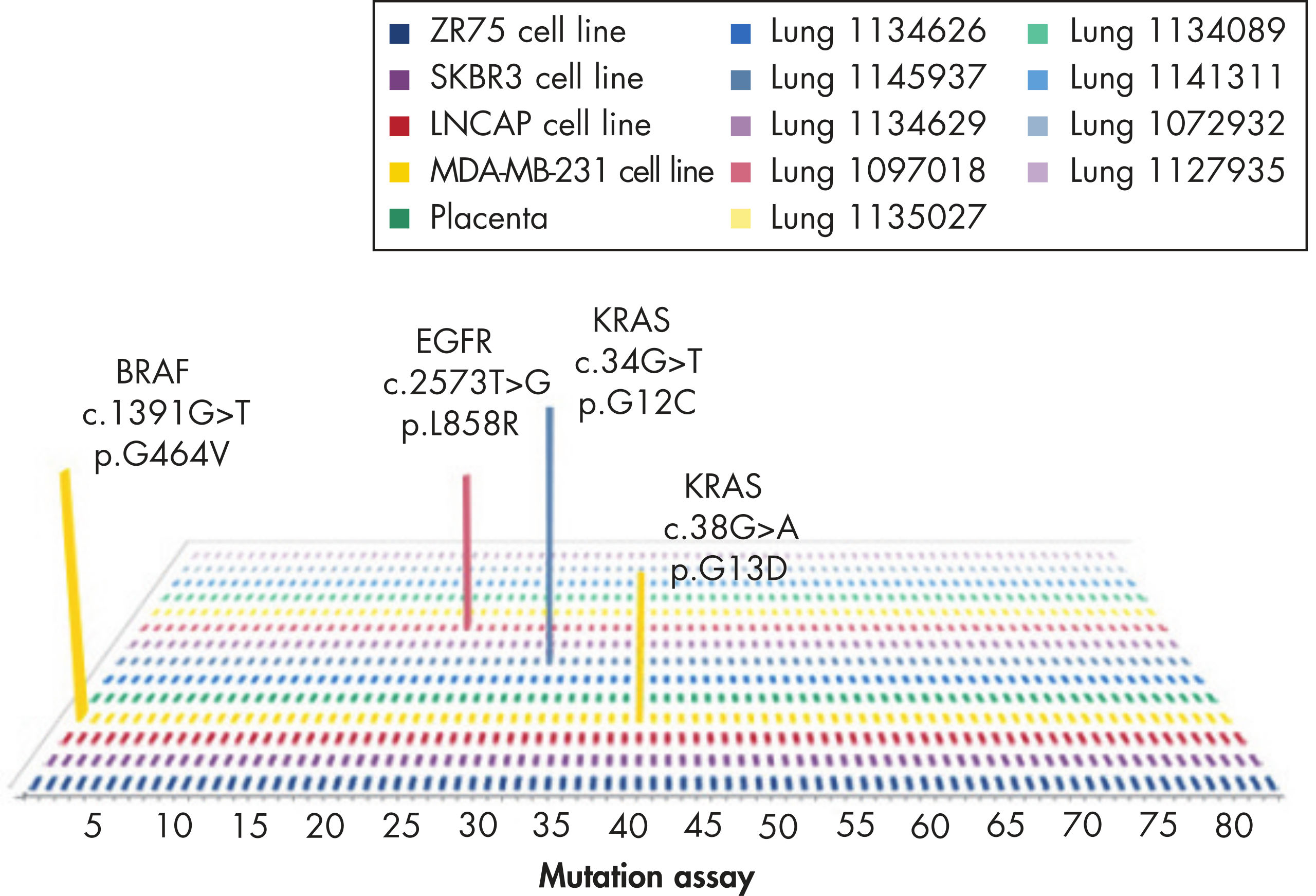
These assays detect the most frequently identified EGFR mutations, which include P-loop and activation loop point mutations, kinase domain deletions, and insertion mutations.
GNAS
Mutations in this gene result in pseudohypoparathyroidism type 1a (PHP1a), which has an atypical autosomal dominant inheritance pattern requiring maternal transmission for full penetrance.
HRAS
The mutation assays detect the most important HRAS variants, all found in codons 12, 13, and 61.
KRAS
The mutation assays detect the most frequently occurring KRAS mutations in codons 12, 13, and 61. Mutations at these positions reduce the protein’s intrinsic GTPase activity and/or cause it to become unresponsive to RasGAP.
MET
The most frequently identified MET gain-of-function point mutations lie in its tyrosine kinase and juxtamembrane domains.
NRAS
The mutation assays detect the most important NRAS variants, all found in codons 12, 13, and 61.
PIK3CA
The most frequently detected PIK3CA gain-of-function mutations occur either in the kinase domain (T1025-G1049, such as H1047L and H1047R) that increase its activity or in the helical domain (P539-E545) which mimic activation by growth factors.
RET
The mutations p.E768D, p.A883F, and p.M918T lie in the protein kinase domain and soluble RET kinase fragment. Most of the remaining mutations lie in predicted extracellular domains.
TSHR
The represented mutations lie in the transmembrane region of this protein and often induce a gain-of-function by increasing basal cAMP levels but could also make the protein slightly less responsive to TSH stimulation. Other mutations lie in either the cytoplasmic or extracellular domains of the protein.