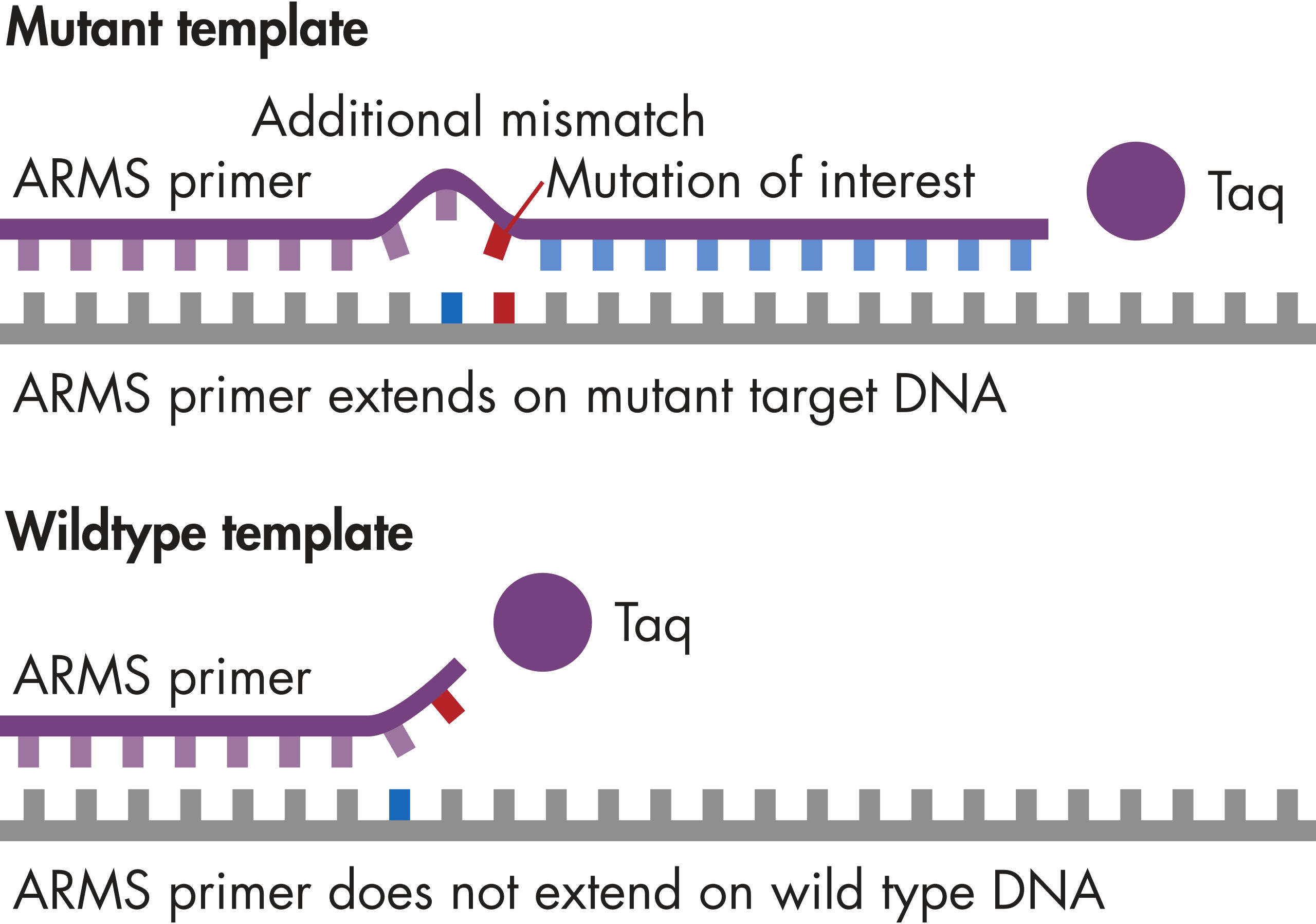
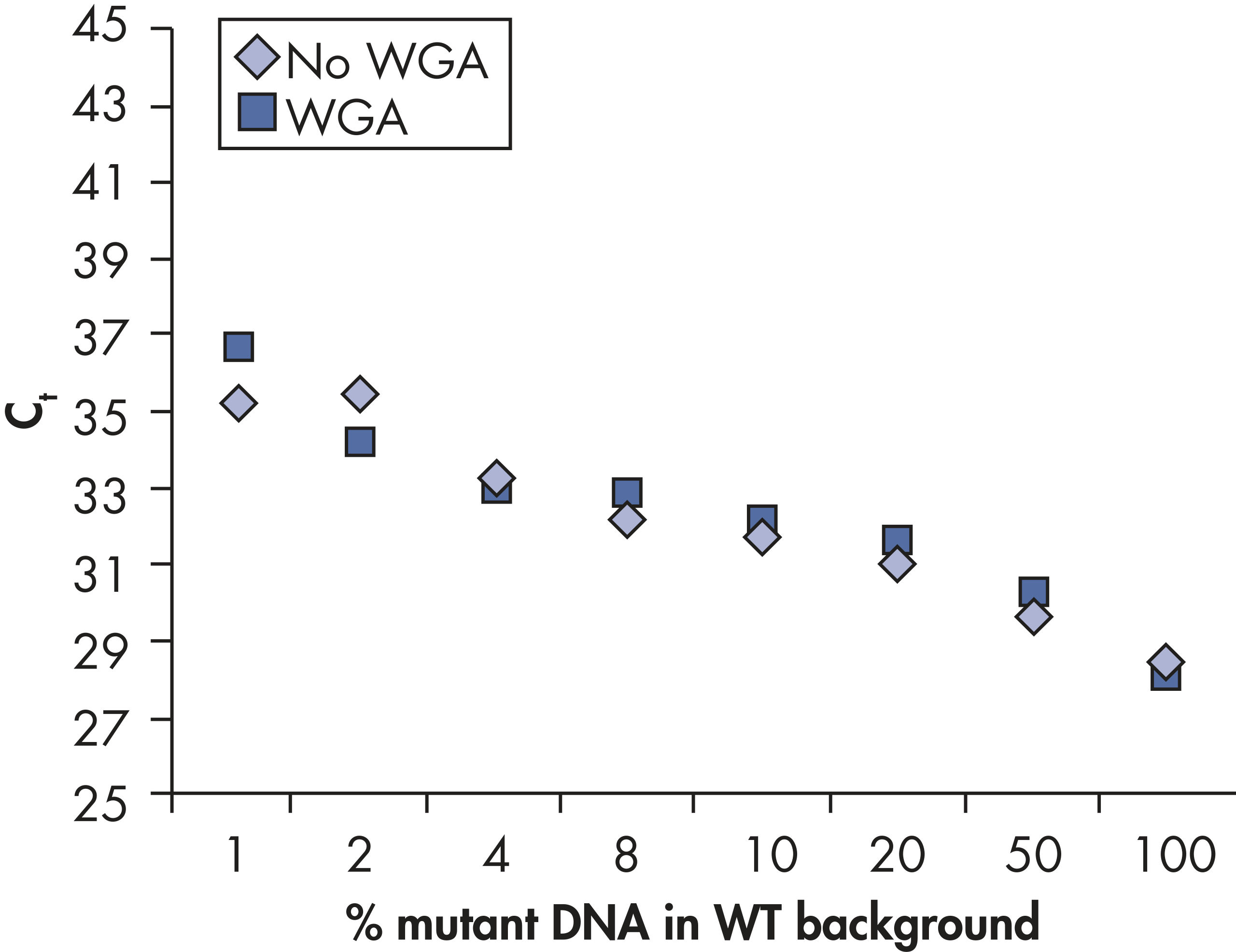
PCR plate and master mix
Target List
AKT1
The best known AKT1 mutations are c.49G>A, p.E17K, a PH domain mutation that results in constitutive targeting of AKT1 to plasma membrane, and an activating mutation, c.145G>A, p.E49K.
BRAF
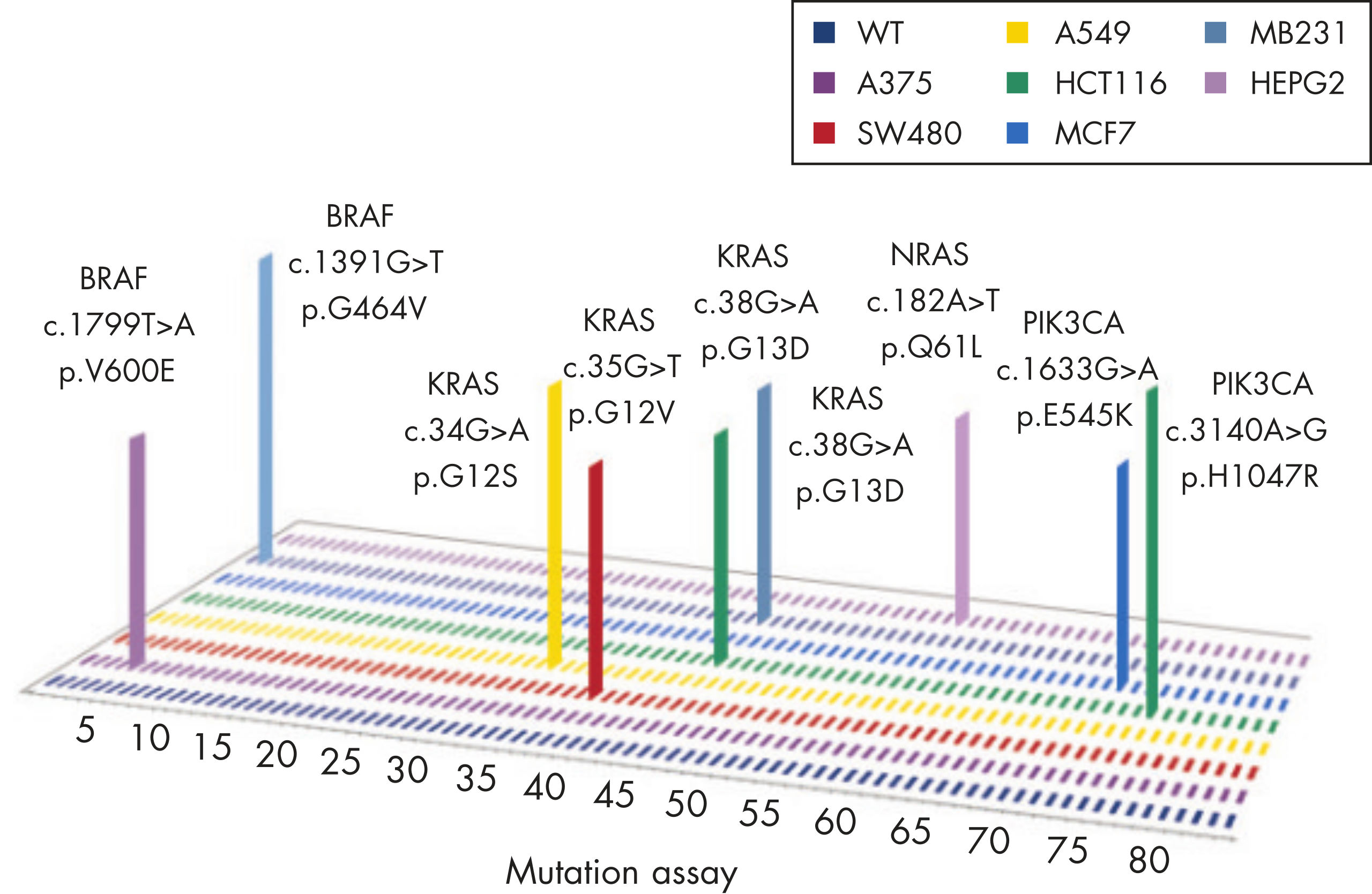
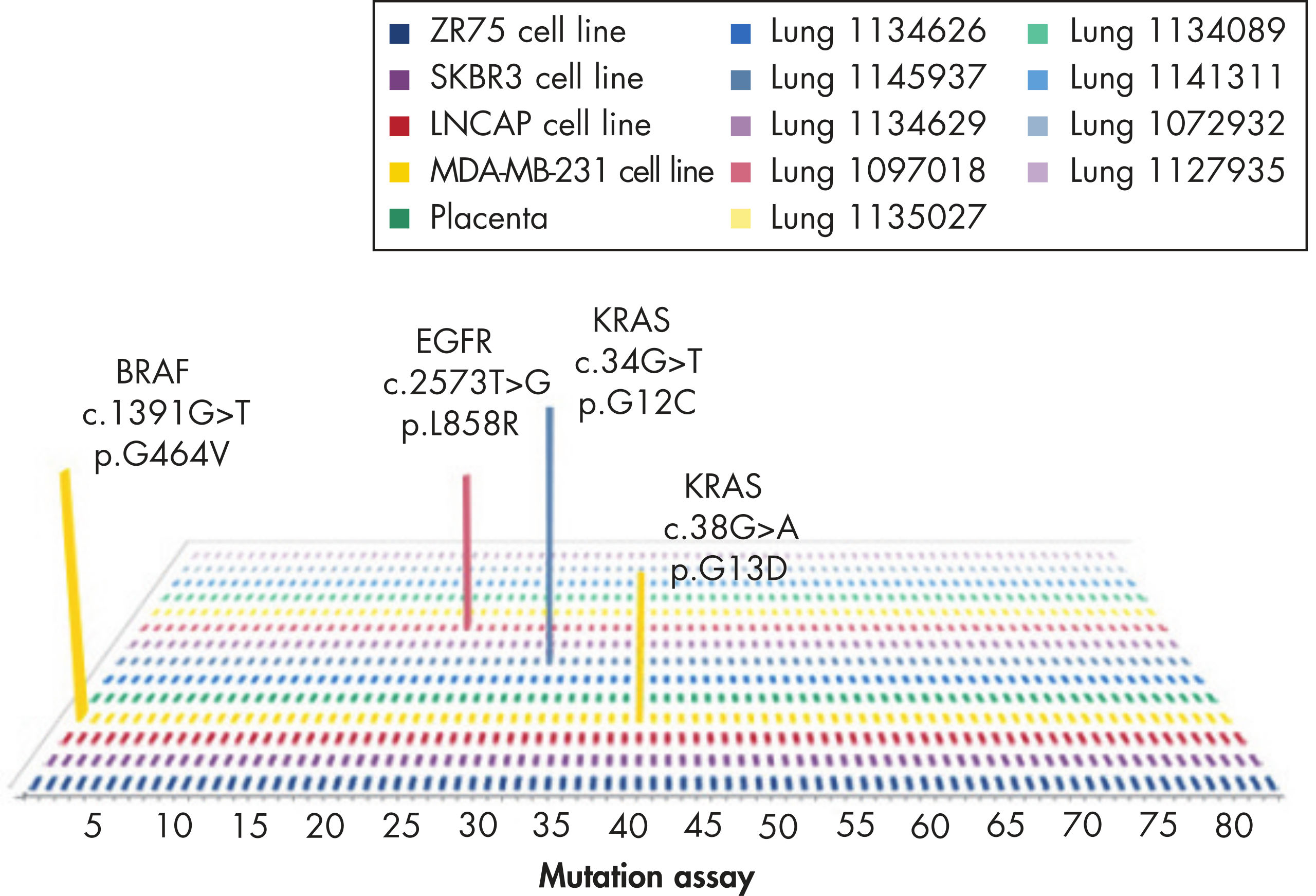
Common point mutations in BRAF either increase kinase activity, such as L597V, L597Q and V600E, or inhibit kinase activity, such as G464V, G466V and G469A.
CTNNB1
The most frequently detected CTNNB1 mutations result in abnormal WNT signaling activity. The mutated codons are mainly several serines and threonines targeted for phosphorylation by GSK3B.
FBXW7
These mutation assays detect FBXW7 variants in either the third or the fourth repeat of the protein's WD40 domain, which is involved in protein-protein interactions.
FGFR2
The most frequently identified FGFR mutations occur in their kinase domains and non-kinase extracellular domains such as the hinge regions and IgG-like domains. Some of the somatic mutations also correspond to congenital SNPs involved in genetic diseases.
HRAS
The mutation assays detect the most important HRAS variants, all found in codons 12, 13, and 61.
KRAS
The mutation assays detect the most frequently occurring KRAS mutations in codons 12, 13, and 61. Mutations at these positions reduce the protein’s intrinsic GTPase activity and/or cause it to become unresponsive to RasGAP.
PIK3CA
The most frequently detected PIK3CA gain-of-function mutations occur either in the kinase domain (T1025-G1049, such as H1047L and H1047R) that increase its activity or in the helical domain (P539-E545) which mimic activation by growth factors.
PTEN
The most commonly detected PTEN loss-of-function mutations either truncate the protein (such as K6fs*4, R130*, R130fs*4, R233*, P248fs*5, and V317fs*3) or cause phosphatase inactivation (such as those at R130 and R173).
TP53
The most frequently detected somatic TP53 mutations occur in the DNA-binding domain which disrupt DNA binding and/or protein structure.